| Aug 2017 Issue 5 | ||||||
 |
||||||
|
|
| Research | |||||||
| A Novel Nanometer-scale Probe Enables a Better Understanding of Cells | |||||||
|
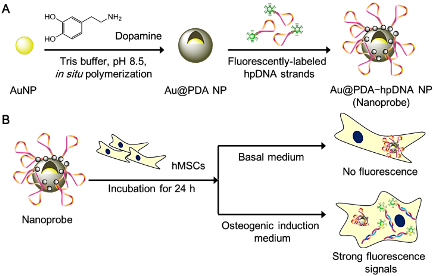
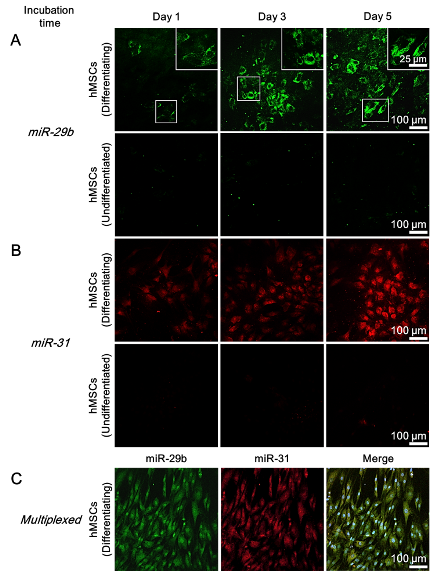
Prof. Liming Bian, Department of Biomedical Engineering MicroRNAs (miRNAs) are single-stranded non-coding RNAs with a typical short length of 21–23 nucleotides.4 They play an important role in controlling the expression of target proteins via either the repression of mRNAs or the inhibition of mRNA translation in a sequence-specific manner, thereby providing an additional level of gene regulation.5,6 In stem cell studies, miRNAs have newly emerged as a mediator of various stem cell behaviors, including differentiation.7-10 miR-29b11 and miR-3112 are two distinct miRNA markers for the osteogenesis of hMSCs. Profiling studies18,21,13 show that these specific miRNAs are significantly up-regulated in stem cells following osteogenic induction. The dynamic nature of these specific miRNA expression profiles mediated by osteogenic differentiation indicates that miRNAs function as viable biomarkers for monitoring the differentiation progress of stem cells. Although much effort has been devoted to tracking intracellular mRNAs,14-17 very few attempts have been reported to detect cancer-related miRNAs in cancerous cell lines.18-19 We now demonstrate that a long-term monitoring (beyond 24 h) of miRNA levels in living stem cells is possible by using the newly designed probe. Existing techniques Conventionally, to determine the differentiation status of hMSCs, end-point methods such as qualitative reverse transcription-polymerase chain reaction (qRT-PCR) and western blot are used. Although these analytical methods are reliable, a large number of cell samples and lysis of cells are required for the analysis, which leads to the unavoidable loss of cell sources. Immunofluorescence and chemical staining are two other common methods to examine the differentiation status of fixed stem cells. However, they are not sensitive enough to monitor the early differentiation stage of stem cells and also preclude real-time monitoring of intracellular activities. Newer techniques including fluorescence-activated cell sorting20,21 (FACS) and surface-enhanced Raman spectroscopy22 (SERS) offer a non-destructive alternative to sort or distinguish between differentiated and undifferentiated stem cells via examination of changes in membranous features in living stem cells. Nevertheless, these techniques generally require expensive staining reagents or specialized instruments and they are not suitable for detecting intracellular biomarkers. Hence, developing a facile and non-invasive way either to monitor the differentiation process or to distinguish the differentiation status of living stem cells was highly desirable. Comparison to exiting techniques Unlike the existing intracellular detection platforms such as Molecular Beacons (MBs), our resultant Au@PDA−hpDNA NPs can naturally enter stem cells without the need of transfection. And different from the NanoFlare, fabrication of our nanoprobes requires only one single type of DNA sequence that can recognize the target miRNA. Due to the close proximity of hpDNAs and the AuNP core (<5 nm)24 and compounded by the intrinsic quenching property of the PDA shell,25 the immobilized hpDNAs on the nanoprobes do not fluoresce appreciably. The presence of two quenching entities (i.e. AuNP core and the PDA shell) makes our nanoprobes unique from the existing nanoparticle-based detection probes that typically possess only a single quenching entity such as the AuNP core of the NanoFlare. In the presence of miRNA targets with a sequence complementary to the recognition region of the immobilized hpDNAs, we show in buffers that the specific binding between the hpDNAs and the target miRNAs triggers the dissociation of the hpDNAs from Au@PDA NPs, and thereafter generates detectable fluorescent signals (Fig. 2). Using these nanoprobes, we demonstrated the specific and long-term detection of two important osteogenic marker miRNAs, namely miR-29b and miR-31, in living hMSCs undergoing osteogenic differentiation as well as living primary osteoblasts (Fig. 2). Summary To summarize, we have demonstrated that our nanoprobes outperform the conventional methods (e.g. qRT-PCR analysis and staining) and the existing commercial intracellular RNA detection probe (e.g. SmartFlareTM) in the context of long-term intracellular detection of miRNAs. More significantly, we have not only established an approach to distinguishing differentiating stem cells from undifferentiated stem cells, but also demonstrated the time-dependent and dynamic expression of specific miRNAs in differentiating stem cells. The capability of our nanoprobes of the multiplexed detection of miRNAs allows enhanced monitoring of cellular events (e.g. differentiation) in living stem cells. More importantly, our nanoprobes afford long-term tracking of intracellular miRNAs in living stem cells which cannot be achieved by commercially available RNA detection probes such as the SmartFlare. The modular design of our nanoprobes offers facile switching of customized hairpin DNA probes, thus opening up an avenue for detecting other biomarkers such as mRNAs in living stem cells. We believe that our Au@PDA−hpDNA nanoprobes show great promise in the investigation of the dynamics of stem cell differentiation, the identification and isolation of specific cell types, and the high-throughput drug screening. 1. Pittenger, M. F.; Mackay, A. M.; Beck, S. C.; Jaiswal, R. K.; Douglas, R.; Mosca, J. D.; Moorman, M. A.; Simonetti, D. W.; Craig, S.; Marshak, D. R., Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284 (5411), 143-7. 2. Abdallah, B. M.; Kassem, M., Human mesenchymal stem cells: from basic biology to clinical applications. Gene Ther 2008, 15 (2), 109-16. 3. Caplan, A. I., Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 2007, 213 (2), 341-7. 4. Stefani, G.; Slack, F. J., Small non-coding RNAs in animal development. Nature reviews. Molecular cell biology 2008, 9 (3), 219-30. 5. Hobert, O., Gene regulation by transcription factors and microRNAs. Science 2008, 319 (5871), 1785-6. 6. Huntzinger, E.; Izaurralde, E., Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics 2011, 12 (2), 99-110. 7. Chen, C. Z.; Li, L.; Lodish, H. F.; Bartel, D. P., MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303 (5654), 83-6. 8. Mizuno, Y.; Yagi, K.; Tokuzawa, Y.; Kanesaki-Yatsuka, Y.; Suda, T.; Katagiri, T.; Fukuda, T.; Maruyama, M.; Okuda, A.; Amemiya, T.; Kondoh, Y.; Tashiro, H.; Okazaki, Y., miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun 2008, 368 (2), 267-72. 9. Ivey, K. N.; Muth, A.; Arnold, J.; King, F. W.; Yeh, R. F.; Fish, J. E.; Hsiao, E. C.; Schwartz, R. J.; Conklin, B. R.; Bernstein, H. S.; Srivastava, D., MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell stem cell 2008, 2 (3), 219-29. 10. Suh, M. R.; Lee, Y.; Kim, J. Y.; Kim, S. K.; Moon, S. H.; Lee, J. Y.; Cha, K. Y.; Chung, H. M.; Yoon, H. S.; Moon, S. Y.; Kim, V. N.; Kim, K. S., Human embryonic stem cells express a unique set of microRNAs. Dev Biol 2004, 270 (2), 488-98. 11. Suh, J. S.; Lee, J. Y.; Choi, Y. S.; Chung, C. P.; Park, Y. J., Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials 2013, 34 (17), 4347-59. 12. Baglio, S. R.; Devescovi, V.; Granchi, D.; Baldini, N., MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013, 527 (1), 321-31. 13. Kaneto, C. M.; Lima, P. S.; Zanette, D. L.; Prata, K. L.; Pina Neto, J. M.; de Paula, F. J.; Silva, W. A., Jr., COL1A1 and miR-29b show lower expression levels during osteoblast differentiation of bone marrow stromal cells from Osteogenesis Imperfecta patients. BMC medical genetics 2014, 15, 45. 14. Seferos, D. S.; Giljohann, D. A.; Hill, H. D.; Prigodich, A. E.; Mirkin, C. A., Nano-flares: probes for transfection and mRNA detection in living cells. Journal of the American Chemical Society 2007, 129 (50), 15477-9. 15. Prigodich, A. E.; Randeria, P. S.; Briley, W. E.; Kim, N. J.; Daniel, W. L.; Giljohann, D. A.; Mirkin, C. A., Multiplexed nanoflares: mRNA detection in live cells. Analytical chemistry 2012, 84 (4), 2062-6. 16. Lin, L. S.; Cong, Z. X.; Cao, J. B.; Ke, K. M.; Peng, Q. L.; Gao, J.; Yang, H. H.; Liu, G.; Chen, X., Multifunctional Fe(3)O(4)@polydopamine core-shell nanocomposites for intracellular mRNA detection and imaging-guided photothermal therapy. ACS nano 2014, 8 (4), 3876-83. 17. Halo, T. L.; McMahon, K. M.; Angeloni, N. L.; Xu, Y.; Wang, W.; Chinen, A. B.; Malin, D.; Strekalova, E.; Cryns, V. L.; Cheng, C.; Mirkin, C. A.; Thaxton, C. S., NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci U S A 2014, 111 (48), 17104-9. 18. Dong, H.; Ding, L.; Yan, F.; Ji, H.; Ju, H., The use of polyethylenimine-grafted graphene nanoribbon for cellular delivery of locked nucleic acid modified molecular beacon for recognition of microRNA. Biomaterials 2011, 32 (15), 3875-82. 19. Ryoo, S. R.; Lee, J.; Yeo, J.; Na, H. K.; Kim, Y. K.; Jang, H.; Lee, J. H.; Han, S. W.; Lee, Y.; Kim, V. N.; Min, D. H., Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS nano 2013, 7 (7), 5882-91. 20. Fukuda, H.; Takahashi, J.; Watanabe, K.; Hayashi, H.; Morizane, A.; Koyanagi, M.; Sasai, Y.; Hashimoto, N., Fluorescence-activated cell sorting-based purification of embryonic stem cell-derived neural precursors averts tumor formation after transplantation. Stem Cells 2006, 24 (3), 763-71. 21. Pruszak, J.; Sonntag, K. C.; Aung, M. H.; Sanchez-Pernaute, R.; Isacson, O., Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells 2007, 25 (9), 2257-68. 22. Kim, T. H.; Lee, K. B.; Choi, J. W., 3D graphene oxide-encapsulated gold nanoparticles to detect neural stem cell differentiation. Biomaterials 2013, 34 (34), 8660-70. 23. Eguchi, A.; Meade, B. R.; Chang, Y. C.; Fredrickson, C. T.; Willert, K.; Puri, N.; Dowdy, S. F., Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nature biotechnology 2009, 27 (6), 567-71. 24. Dubertret, B.; Calame, M.; Libchaber, A. J., Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nature biotechnology 2001, 19 (4), 365-70. 25. Qiang, W.; Li, W.; Li, X.; Chen, X.; Xu, D., Bioinspired polydopamine nanospheres: a superquencher for fluorescence sensing of biomolecules. Chemical Science 2014, 5 (8), 3018-3024.
|
|
||||||
|
|
|||||||
 |
|||||||
|
|
|||||||||||||||||||||||